Multithreading
Multithreading is part of a free web series, ChemPlugin Modeling with Python, by Aqueous Solutions LLC.
What you need:
- ChemPlugin SDK 2025 recommended
-
Input files:
 RTM1.cpp,
RTM1.cpp,  RTM2.cpp
RTM2.cpp
Download this unit to use in your courses:
- PowerPoint slides (.pptx)
Click on a file to open, or right-click and select “Save link as…” to download.
Introduction
Modern personal computers commonly contain more than one computing core. A multi-threaded application gains a performance advantage by running on more than one core at the same time.
Although Python is not set up well for multithreading applications, we'd like to discuss how client programs written in other langues, such as C++, can implement ChemPlugin objects to run in parallel.
To multithread a client on a conventional computer, a programmer likely works with the OpenMP application programmer interface (API). In our discussion here, we use OpenMP to multithread the “RTM1.cpp” client, the C++ equivalent of the Python script we discussed in the previous lesson.
The diagram below represents a multi-core processor
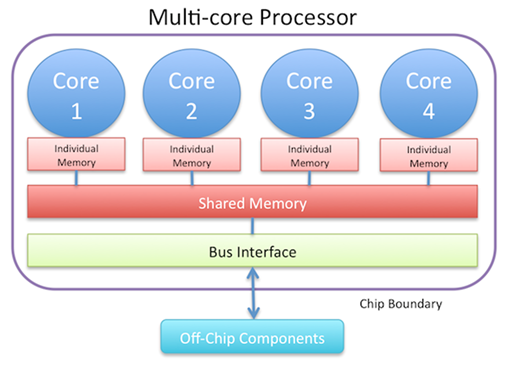
A multithreaded application runs threads of a process on more than one core. A hyperthreaded app runs two threads per core. The basic steps in multithreading a client program are shown below.
First, include the header file:
#include <omp.h>
Note the hash (“#”) symbols in C++ are not for commented lines, as in Python. Rather, they indicate preprocessor commands. File “omp.h” is the C++ header for OpenMP.
Next, set the compiler option
-Qopenmp
The “-Qopenmp” keyword is required to produce a multithreaded executable.
Next, prepend to loops
#pragma omp parallel for
The statements following the “#pragma” directive split creation of the ChemPlugin instances across a work-sharing loop.
Finally, set lots of instances! In general, creating a parallel region in a client program required the system to expend overhead. If there's too little work to share, the client may spend as much time administering the parallel region as it gains from the work sharing. As a result, it's best to set lots of instances to take good advantage of multithreading the client.
An example of a work-sharing loop is shown below:
#pragma omp parallel for
for (int i=0; i<nx; i++)
cp[i]->AdvanceChemical();
Instantiation
Instantiation of a ChemPlugin instance involves a non-trivial amount of work. Each instance, upon being created, lays out memory for itself, reads in a thermodynamic dataset, and prepares itself to accept configuration commands. Significant improvement to the time required for an application to start up can be gained by multithreading the instantiation step.
The original instantiation
ChemPlugin *cp = new ChemPlugin[nx];
... some code ...
cp[i].Config(cmd);
is strictly serial; a single thread lays out one instance after another until “nx” isntances have been created.
One way to multithread the initialization is to cast “cp” as a vector of pointers to ChemPlugin instances, instead of a vector of the instances themselves. In this way, we can instantiate in parallel:
std::vector<ChemPlugin*> cp(nx);
#pragma omp parallel for
for (int i=0; i<nx; i++)
cp[i] = new ChemPlugin();
... some code ...
cp[i]->Config(cmd);
Here, the statements following the “#pragma” directive split creation of the ChemPlugin instances across a work-sharing loop.
Each vector element “cp[i]” is a pointer to a ChemPlugin instance now, rather than a reference to an instance itself. Hence, we need to change constructions like
cp[i].Config(cmd);
throughout “RTM1.cpp” to
cp[i]->Config(cmd);
Reduction
A serial loop can be broken :
for (int i=0; i<nx; i++)
if (cp[i]->AdvanceChemical())
end_run(-1);
Work-sharing loops in OpenMP cannot be broken within the parallel region, however. To parallelize the loop, use a reduction variable “nerror” to count the number of times the loop encounters a condition that would cause it to break:
int nerror = 0;
#pragma omp parallel for reduction(+ : nerror)
for (int i=0; i<nx; i++)
if (cp[i]-> AdvanceChemical())
nerror++;
if (nerror) end_run(-1);
If “nerror” is non-zero after the loop completes, the client exits.
Need for privacy
The loop over which client “RTM1.cpp” links the ChemPlugin instances
CpiLink link;
for (int i=1; i<nx; i++) {
link = cp[i].Link(cp[i-1]);
link.FlowRate(flow);
}
cannot work correctly as listed, because within the parallel region the various threads would write to and read from “link” simultaneously.
Instead, “link” must be re-declared within the scope of the loop
#pragma omp parallel for
for (int i=1; i<nx; i++) {
CpiLink link = cp[i].Link(cp[i-1]);
link.FlowRate(flow);
}
Inside the parallel region, now, “link” is private to each thread.
Speedup
To test the extent to which multithreading the client program “RTM2.cpp” sped its execution relative to the single-threaded version of “RTM1.cpp”, we timed the solution of three problems using varying numbers “nx” of ChemPlugin instances. In each case, we report speedup as the clock time required to solve the problem using the single-threaded relative to the multithreaded client.
The speedups observed on a quad core processor for the multithreaded client on runs made using 4 000, 10 000, and 40 000 ChemPlugin instances are:
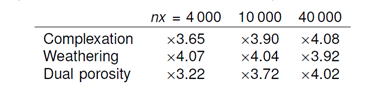
The nominal maximum speedup on a quad core processor is x4. It is possible, as we see in this table, to exceed this limit somewhat by hyperthreading—i.e, scheduling two threads on each core.
Authors
Craig M. Bethke and Brian Farrell. © Copyright 2016–2026 Aqueous Solutions LLC. This lesson may be reproduced and modified freely to support any licensed use of The Geochemist's Workbench® software, provided that any derived materials acknowledge original authorship.
References
Bethke, C.M., 2022, Geochemical and Biogeochemical Reaction Modeling, 3rd ed. Cambridge University Press, New York, 520 pp.
Bethke, C.M., 2026, The Geochemist's Workbench®, Release 18: ChemPlugin™ User's Guide. Aqueous Solutions LLC, Champaign, IL, 303 pp.
Comfortable with multithreading?
Return to the ChemPlugin Modeling with Python home.